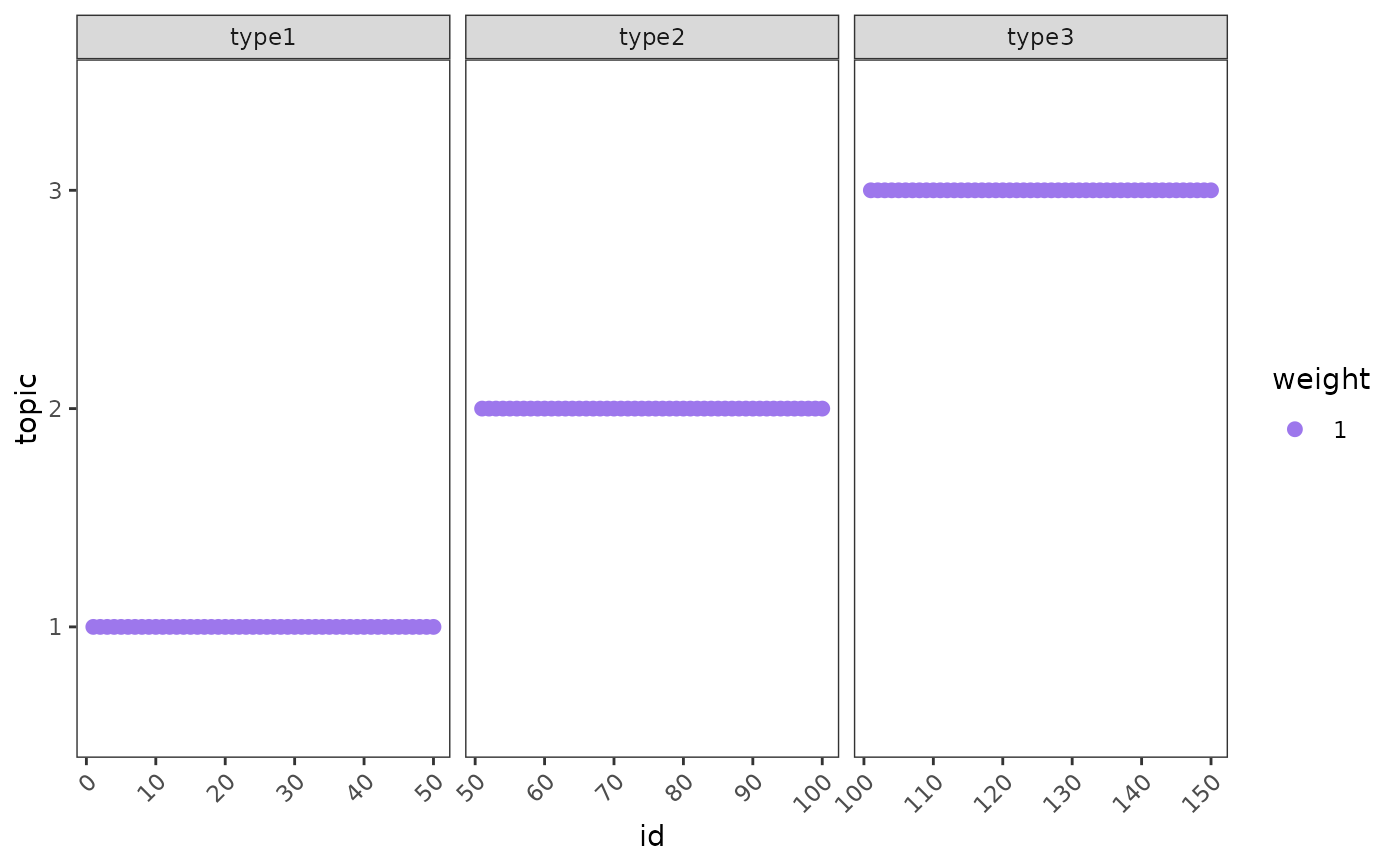
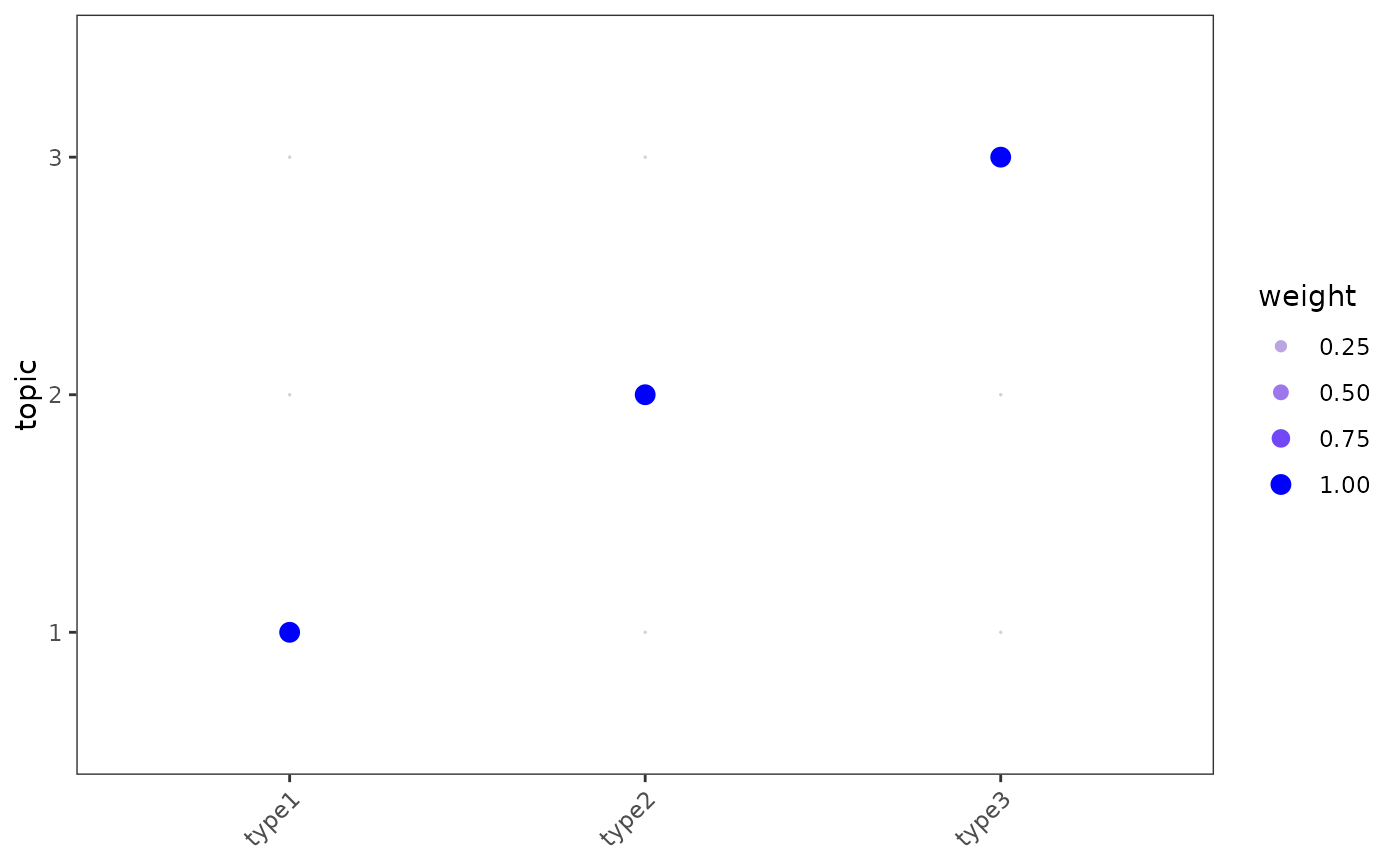
This function takes in the fitted NMF model and returns the
topic profiles learned for each cell facet = FALSE or cell type
facet = TRUE. Ideal training will return all the cell from the same
cell type to share a unique topic profile.
Usage
# S4 method for NMF,ANY
plotTopicProfiles(x, y, ...)
# S4 method for NMF,character
plotTopicProfiles(x, y, facet = FALSE, min_prop = 0.1, ncol = NULL)Arguments
- x
NMFfitobject- y
vector of group labels. Should be of length
ncol(coef(x)).- ...
additional parameters
- facet
logical indicating whether to stratify by group. If
FALSE(default), weights will be the median across cells for each group (point = topic weight for a given cell type). IfTRUE, cell-specific weights will be shown (point = topic weight of a given cell).- min_prop
scalar in [0,1]. When
facet = TRUE, only cells with a weight >min_propwill be included.- ncol
integer scalar specifying the number of facet columns.